Translation Notice
This article was machine-translated using DeepSeek-R1.
- Original Version: Authored in Chinese by myself
- Accuracy Advisory: Potential discrepancies may exist between translations
- Precedence: The Chinese text shall prevail in case of ambiguity
- Feedback: Technical suggestions regarding translation quality are welcomed
Preface
We often encounter problems like this in real life:
Highway Construction
There are several cities where highways need to be built between them. Each pair of cities can be connected via a bidirectional road. Each road between two cities has a specific construction cost. How can we construct roads with the minimal total cost while ensuring all cities are connected directly or indirectly?
We can abstract this problem by representing cities as graph vertices and roads as weighted undirected edges, forming a weighted undirected graph. Since we want minimal total cost, the constructed roads must form a spanning tree, transforming the problem into:
Given a weighted undirected graph $G$, find its spanning tree where the sum of all edge weights is minimized.
This is the well-known Minimum Spanning Tree (MST) problem.
Consider the following graph:
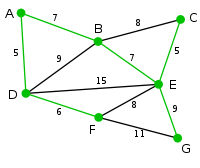
The green edges form its minimum spanning tree.
Mathematicians have studied this problem extensively. However, focusing only on correctness without considering simplicity, feasibility, and efficiency led many algorithms to obsolescence. Two algorithms stood out: Kruskal and Prim.
Template Problem: Luogu P3366 【Template】Minimum Spanning Tree
Data Constraints: $N\le5000,M\le2\times10^5,w\le 10^4$.
Kruskal
The Kruskal algorithm, proposed by Joseph Kruskal in 1956, has a time complexity of $\mathcal O(m\log m)$. Let’s examine its workflow.
Kruskal Algorithm Workflow
- Sort all edges in ascending order of weight and iterate through them.
- For each edge, if adding it to the current subgraph doesn’t form a cycle, include it as part of the MST.
- The algorithm terminates after selecting $N-1$ edges.
Disjoint-Set Union (DSU) — Accelerating the Algorithm
Before implementing Kruskal, we need DSU. Using DFS for cycle detection would result in $\mathcal O(nm + m\log m)$ time complexity, which is inefficient. DSU reduces this to $\mathcal O(m\log m)$ (dominated by sorting). DSU Template:
|
|
Using DSU, the algorithm’s time complexity becomes $\mathcal O(m\log m)$. Now, let’s see the code.
Reference Code
For readers unfamiliar with DSU, feel free to copy the template first.Study it later
For single Kruskal runs, use priority_queue (more efficient than sort). For multiple runs, pre-sort edges.
|
|
Alternative ending (slower but concise):
|
|
Prim
The Prim algorithm was discovered by Czech mathematician Vojtěch Jarník in 1930, independently by Robert C. Prim in 1957, and rediscovered by Edsger W. Dijkstra in 1959. Hence, it’s also called DJP, Jarník, or Prim-Jarník algorithm.
Prim Algorithm Workflow
Similar to Dijkstra, vertices are divided into sets $S$ and $T$:
- Initially, all vertices are in $S$, $T$ is empty.
- Move any vertex from $S$ to $T$.
- Repeat until all vertices are in $T$:
- Select edge $(u,v,w)$ where $u\in S$, $v\in T$, with minimal $w$.
- Add this edge to MST and move $u$ to $T$.
We present optimized versions using priority_queue and set.
Priority Queue Optimization
Runtime: $328\mathrm{ms}$
Time Complexity: $\mathcal O(n\log m)$
|
|
Set Optimization
Runtime: $351\mathrm{ms}$
Time Complexity: $\mathcal O(n\log n)$
|
|
Exercises
- Luogu P1551 Relatives (DSU practice)
- UVA1151 Buy or Build (Kruskal)
- UVA1395 Slim Span (Kruskal)
- Luogu P1265 Highway Construction (Prim)
Summary
Comparison of Kruskal and Prim:
| Metric | Kruskal | Prim |
|---|---|---|
| Time Complexity | $\mathcal O(m\log m)$ | $\mathcal O(n\log m)$1 |
| Runtime2 | $255\mathrm{ms}$ | $328\mathrm{ms}$ |
| Code Complexity | Low | Medium |
| Applicability | Sparse Graphs | Dense Graphs |
In most cases, Kruskal is preferred. Use Prim for specific requirements.
If you find this article helpful, please like/share/bookmark. Thanks for your support!
-
For priority_queue optimization. Set optimization is $\mathcal O(n\log n)$. ↩︎
-
Results from Luogu P3366. Kruskal uses DSU+priority_queue; Prim uses priority_queue. ↩︎